FundRmentals 08
Dr Danielle Evans | University of Sussex
Setup & Q&A
Open RStudio
Open/create your fundRmentals R Project (click the blue cube in the top right corner of RStudio)
Open a new Rmd file for today
If you missed last week, install the palmerpenguins package - install.packages("palmerpenguins")
Install the GGally, correlation, & effectsize packages
In a new code chunk, load tidyverse, palmerpenguins, GGally, correlation & effectsize using the library() command
In another code chunk, create peng_data by copying the below code:
peng_data <- palmerpenguins::penguins %>% na.omit()Any questions from the last tutorial?
Overview
Correlations
Independent t-test
Reporting results with inline code (extRa)
Next steps
Correlations: Visualization
Visualising relationships is made easy with the GGally::ggscatmat() function
GGally::ggscatmat() shows us a correlation matrix, with distributions, scatterplots, & correlation coefficients
To use it, we give the names of our data & the variables we want to examine
GGally::ggscatmat(data, columns = c("variable 1", "variable 2", "variable 3"))
Task: using peng_data, use the GGally::ggscatmat() function on body_mass_g, flipper_length_mm, bill_depth_mm
GGally::ggscatmat(peng_data, columns = c("body_mass_g", "flipper_length_mm", "bill_depth_mm"))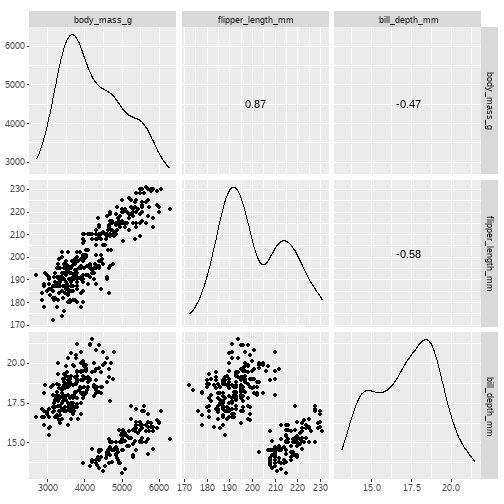
Correlations: Tests
- We can use the correlation::correlation() function to perform a correlation, the default options are given below:
correlation::correlation(data, method = "pearson", p_adjust = "holm", ci = 0.95 )- If we are happy with these defaults, we can pipe in the data we want to use, and select the variables:
data %>% dplyr::select(variable_1, variable_2) %>% correlation::correlation()Task: perform a correlation with our peng_data, on the body_mass_g & flipper_length_mm variables, save it in an object (<-)
p_adjust. By default the function corrects the p-value for the number of tests you have performed (a good idea) using the Holm-Bonferroni method, which applies the Bonferroni criterion in a slightly less strict way that controls the Type I error rate but with less risk of a Type II error. You can change this argument to none (i.e. don’t correct for multiple tests, a bad idea), bonferroni (to apply the standard Bonferroni method) or several other methods.
Correlations: Tests
peng_cor <- peng_data %>% dplyr::select(body_mass_g, flipper_length_mm) %>% correlation::correlation()peng_cor## # Correlation Matrix (pearson-method)## ## Parameter1 | Parameter2 | r | 95% CI | t(331) | p## --------------------------------------------------------------------------## body_mass_g | flipper_length_mm | 0.87 | [0.84, 0.90] | 32.56 | < .001***## ## p-value adjustment method: Holm (1979)## Observations: 333We can even edit the number of decimal places if we wanted to:
peng_cor <- peng_data %>% dplyr::select(body_mass_g, flipper_length_mm) %>% correlation::correlation(digits = 3, ci_digits = 3)Comparing Independent Means: t-test
We can use the t.test function to perform a t-test, this function has the following arguments & defaults:
We sub in our column names for the outcome & predictor, the name of our dataset, whether we want a paired t-test or not, whether we want Welch's correction (the correction is the default), the confidence interval, and whether to exclude missing data (na.action = na.exclude)
t.test(outcome ~ binary_predictor, data = data, paired = FALSE, var.equal = FALSE, conf.level = 0.95)
Task: perform an independent t-test with our peng_data, using the sex & body_mass_g variables, keep the default options of Welch's correction & 95% CIs, give the object a name (<-)
Comparing Independent Means: t-test
body_mass_test <- t.test(body_mass_g ~ sex, data = peng_data)body_mass_test## ## Welch Two Sample t-test## ## data: body_mass_g by sex## t = -8.5545, df = 323.9, p-value = 4.794e-16## alternative hypothesis: true difference in means is not equal to 0## 95 percent confidence interval:## -840.5783 -526.2453## sample estimates:## mean in group female mean in group male ## 3862.273 4545.685Comparing Independent Means: Effect Size
- We can calculate effect size just as easy as doing a t-test with the effectsize::cohens_d() function:
cohens_d <- effectsize::cohens_d(outcome ~ binary_predictor, data = data)Task: calculate cohen's d for the t-test we just did! Make sure to save it in an object
Comparing Independent Means: Effect Size
- We can calculate effect size just as easy as doing a t-test with the effectsize::cohens_d() function:
cohens_d <- effectsize::cohens_d(outcome ~ binary_predictor, data = data)Task: calculate cohen's d for the t-test we just did! Make sure to save it in an object
cohens_d <- effectsize::cohens_d(body_mass_g ~ sex, data = peng_data)cohens_d## Cohen's d | 95% CI## --------------------------## -0.94 | [-1.16, -0.71]## ## - Estimated using pooled SD.ExtRa: Reporting Results
We can report the results of these tests using inline code:
By saving all our results into objects we can access the values contained within them

We need to know the name of the object & the name of the element
We can look in our environment pane & click the blue 'play button' next to the name of the object to see what it contains & what the elements are called
We can then select individual elements by using the $
If there's more than one value in an element, we can use [] & the number of the one we want
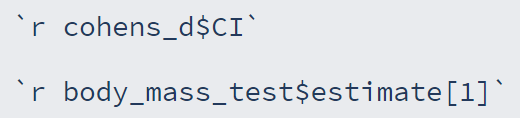
Task: try it out on some of your results & knit your doc!
Next Steps
Save the RMarkdown document from today Ctrl + S or Command + S
For next week you should complete the discovr_08 tutorial on the linear model by pasting the below code into the Console in RStudio, pressing Enter
learnr::run_tutorial("discovr_08", package = "discovr")write_csv(peng_cor, file = "cor.csv") write_csv(broom::tidy(body_mass_test), file = "ttest.csv") write_csv(cohens_d, file = "cohens.csv")